Revolutionary Discovery: DNA’s New Frontier
DNA DNA, or Deoxyribonucleic Acid, is the genetic material found in cells, composed of a double helix structure. It serves as the genetic blueprint for all living organisms. Read Full Definition, the cornerstone of genetic material, traditionally comprises four nucleobases: adenine (A), thymine (T), guanine (G), and cytosine (C). These nucleobases form the genetic code in all living organisms. However, a groundbreaking discovery in the bacteriophage S-2L has revealed an alternative nucleobase, 2-aminoadenine (Z), which replaces adenine and pairs with thymine (Z–T) through three hydrogen bonds instead of the conventional two.
DNA, or Deoxyribonucleic Acid, is the genetic material found in cells, composed of a double helix structure. It serves as the genetic blueprint for all living organisms. Read Full Definition, the cornerstone of genetic material, traditionally comprises four nucleobases: adenine (A), thymine (T), guanine (G), and cytosine (C). These nucleobases form the genetic code in all living organisms. However, a groundbreaking discovery in the bacteriophage S-2L has revealed an alternative nucleobase, 2-aminoadenine (Z), which replaces adenine and pairs with thymine (Z–T) through three hydrogen bonds instead of the conventional two.
Cyanophage S-2L: An Exception to the Genetic Rule
Cyanophage S-2L is a virus that infects bacteria and is unique in its DNA composition. The incorporation of Z instead of A provides the phage’s DNA with exceptional stability, particularly at elevated temperatures. This structural alteration also makes the DNA less recognizable by typical proteins and molecules, possibly conferring an evolutionary advantage by enhancing the phage’s resistance to environmental stressors.
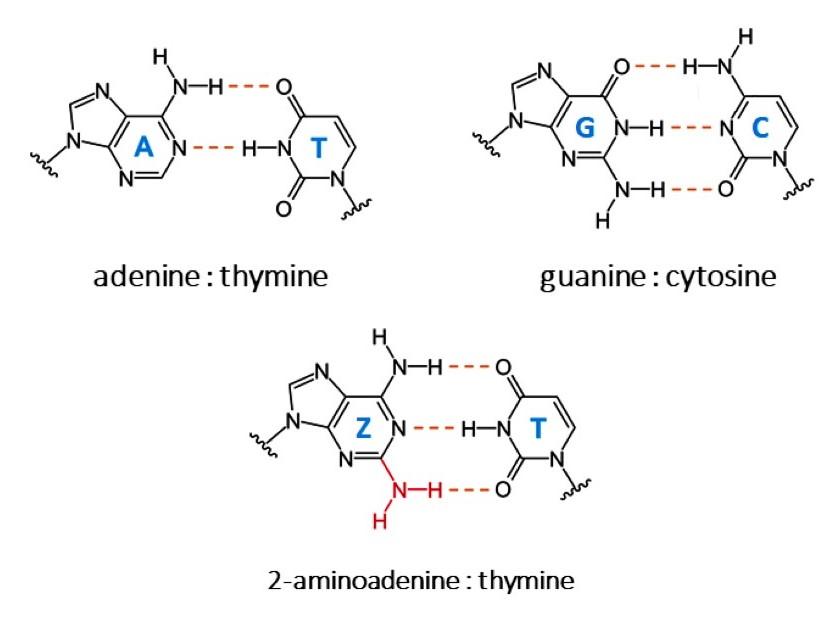
A : T, G : C and Z : T bonds © Dona Sleiman et al.
Unveiling the Biosynthesis Pathway of 2-Aminoadenine
Since its identification in 1977, the biosynthetic origin of the Z base remained a mystery. Researchers at the Institut Pasteur and CNRS, in collaboration with CEA, have now uncovered the enzymatic pathway responsible for the production of 2-aminoadenine.
Key findings include:
- Identification of the PurZ Enzyme: A homolog of succinoadenylate synthase (PurA), named PurZ, was discovered in the genome

- Ancient Origins: Phylogenetic analysis linked PurZ to archaea, suggesting it is an ancient enzyme that may have offered evolutionary benefits.
- Crystallographic ValidationValidation, often referred to as method validation, is a crucial process in the laboratory when introducing a new machine, technology, or analytical technique. It involves a series of systematic steps and assessments to ensure that Read Full Definition: The structural analysis conducted at the Institut Pasteur’s Crystallography Platform confirmed the enzymatic mechanisms involved in synthesizing the Z nucleobase.
Implications for Genetics and Biotechnology
The discovery of the Z–T base pair and its biosynthetic pathway demonstrates the feasibility of incorporating new bases into genetic material enzymatically. This breakthrough expands the potential coding capacityThe amount of finished product that could be produced, either in one batch or over a defined period of time, and given a set list of variables. Read Full Definition of DNA and opens new avenues in synthetic biology. Scientists can now explore creating synthetic genetic biopolymers with enhanced stability and functionality, which may have applications in fields like medicine, data Information in analog or digital form that can be transmitted or processed. Read Full Definition storage, and biotechnology.
Information in analog or digital form that can be transmitted or processed. Read Full Definition storage, and biotechnology.
Future Prospects: Redefining Genetic Boundaries
This discovery challenges the notion of DNA’s four-base limitation and paves the way for the development of synthetic genomes. By harnessing the stability and unique properties of the Z base, researchers could design robust genetic systems tailored for specific purposes, such as high-temperature environments or advanced molecular computing.
Journal Reference
Dona Sleiman, Pierre Simon Garcia, Marion Lagune, Jerome Loc’h, Ahmed Haouz, Najwa Taib, Pascal Röthlisberger, Simonetta Gribaldo, Philippe Marlière, Pierre Alexandre Kaminski. “A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.” Science, 2021; 372 (6541): 516. DOI: 10.1126/science.abe6494